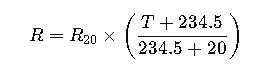Understanding Dose Calculations for Restriction Enzymes, Ligases, and Polymerases
Precise dose calculation is critical for enzymatic reactions in molecular biology workflows. It ensures optimal activity and reproducibility.
This article details formulas, tables, and real-world examples for calculating doses of restriction enzymes, ligases, polymerases, and related reagents.
- Calculate the units of restriction enzyme needed for digesting 1 µg of DNA in 50 µL reaction volume.
- Determine the amount of T4 DNA ligase required to ligate 100 ng of vector and insert at a 1:3 molar ratio.
- Calculate the volume of polymerase to add for a 25 µL PCR reaction with 10 ng template DNA.
- Estimate the units of alkaline phosphatase needed to dephosphorylate 500 ng of linearized plasmid DNA.
Comprehensive Tables of Common Enzymatic Dose Parameters
| Enzyme Type | Typical Unit Definition | Recommended Units per Reaction | Typical Reaction Volume (µL) | Optimal Temperature (°C) | Common Buffer |
|---|---|---|---|---|---|
| Restriction Enzymes | 1 Unit = 1 µg DNA digested in 1 hour at 37°C | 5–10 Units | 20–50 | 37 | NEBuffer 2.1, CutSmart |
| T4 DNA Ligase | 1 Unit = ligates 50% of 100 ng DNA in 30 min at 25°C | 1–3 Units | 10–20 | 16–25 | Ligase Buffer (ATP-containing) |
| Taq DNA Polymerase | 1 Unit = incorporates 10 nmol dNTP into acid-insoluble material in 30 min at 74°C | 0.5–2.5 Units | 20–50 | 72 | PCR Buffer with MgCl2 |
| Alkaline Phosphatase (CIP) | 1 Unit = removes 1 nmol phosphate/min at 37°C | 1–5 Units | 10–50 | 37 | NEBuffer 3.1 |
| Phusion High-Fidelity Polymerase | 1 Unit = incorporates 10 nmol dNTP in 30 min at 72°C | 0.5–1 Unit | 20–50 | 72 | Phusion HF Buffer |
| DNA Polymerase I (Klenow Fragment) | 1 Unit = incorporates 10 nmol dNTP in 30 min at 37°C | 1–5 Units | 20–50 | 37 | Klenow Buffer |
| RNAse H | 1 Unit = hydrolyzes 1 nmol RNA in RNA-DNA hybrid/min at 37°C | 1–3 Units | 20–50 | 37 | RNAse H Buffer |
| DNA Ligase (E. coli) | 1 Unit = ligates 1 nmol of DNA ends/min at 25°C | 1–5 Units | 10–50 | 25 | E. coli Ligase Buffer |
Fundamental Formulas for Dose Calculations
Calculating the correct enzyme dose requires understanding the relationship between enzyme units, substrate amount, reaction volume, and incubation time. Below are the key formulas and detailed explanations of each variable.
1. Restriction Enzyme Dose Calculation
The general formula to calculate the units of restriction enzyme required is:
- DNA amount (µg): The mass of DNA to be digested.
- Units per µg DNA: Typically 1 Unit digests 1 µg DNA in 1 hour under optimal conditions.
- Adjustment factor: Accounts for reaction time, enzyme efficiency, and buffer conditions (usually 1–2).
For example, if digesting 2 µg DNA in 1 hour, and using 1 Unit per µg DNA, the enzyme units required would be:
2. DNA Ligase Dose Calculation
Ligase dosing depends on the molar ratio of insert to vector and total DNA amount. The formula to calculate ligase units is:
- Total DNA amount (ng): Sum of vector and insert DNA.
- Units per 100 ng DNA: Typically 1 Unit ligates 100 ng DNA in 30 minutes.
- Adjustment factor: Usually 1–3 depending on ligation efficiency and reaction conditions.
For example, ligating 300 ng total DNA with 1 Unit per 100 ng and an adjustment factor of 2:
3. Polymerase Dose Calculation for PCR
Polymerase units are often standardized per reaction volume and template amount. The formula is:
- Reaction volume (µL): Total PCR reaction volume.
- Standard volume (µL): Usually 50 µL.
- Standard units: Typically 1–2 Units per 50 µL reaction.
- Adjustment factor: Adjusts for template complexity or desired fidelity (1–2).
For a 25 µL reaction using 1 Unit per 50 µL standard and adjustment factor 1.5:
4. Alkaline Phosphatase Dose Calculation
Alkaline phosphatase dosing depends on DNA amount and reaction time:
- DNA amount (ng): Amount of DNA to be dephosphorylated.
- Units per 100 ng DNA: Typically 1 Unit per 100 ng DNA.
- Adjustment factor: Usually 1–3 depending on incubation time and enzyme activity.
For 500 ng DNA with 1 Unit per 100 ng and adjustment factor 2:
Detailed Explanation of Variables and Common Values
- Unit Definition: Enzyme activity is standardized by manufacturers. For restriction enzymes, 1 Unit typically digests 1 µg DNA in 1 hour at optimal temperature.
- Adjustment Factor: Accounts for suboptimal conditions, incomplete digestion, or shorter incubation times. Usually ranges from 1 to 3.
- Reaction Volume: Enzyme concentration and buffer composition depend on total reaction volume, which typically ranges from 10 to 50 µL.
- DNA Amount: Accurate quantification of DNA is essential. Overloading or underloading affects enzyme efficiency.
- Temperature: Each enzyme has an optimal temperature for activity, e.g., 37°C for most restriction enzymes, 16–25°C for ligases, and 72°C for polymerases.
- Buffer Composition: Buffers provide optimal ionic strength, pH, and cofactors (e.g., Mg2+, ATP) necessary for enzyme function.
Real-World Application Examples
Example 1: Restriction Enzyme Digestion of Plasmid DNA
A molecular biologist needs to digest 3 µg of plasmid DNA with EcoRI in a 50 µL reaction. The manufacturer states 1 Unit digests 1 µg DNA in 1 hour at 37°C. The digestion will be performed for 45 minutes. Calculate the units of EcoRI required.
Step 1: Calculate base units for 3 µg DNA:
Step 2: Adjust for shorter incubation time (45 min instead of 60 min). Adjustment factor = 60/45 = 1.33
Step 3: Calculate total units:
Result: Add 4 Units of EcoRI to the 50 µL reaction to ensure complete digestion in 45 minutes.
Example 2: Ligation of Insert into Vector
A researcher wants to ligate a 500 ng vector with a 1.5 kb insert at a 1:3 molar ratio. The vector is 3 kb in size. The ligation reaction volume is 20 µL. The ligase activity is 1 Unit per 100 ng DNA in 30 minutes at 16°C. Calculate the units of T4 DNA ligase required.
Step 1: Calculate molar amounts:
- Vector size = 3,000 bp
- Insert size = 1,500 bp
- Vector mass = 500 ng
Molar amount of vector:
= 500 / (3000 × 660) × 10-9
= 500 / 1,980,000 × 10-9
≈ 2.53 × 10-13 moles
Insert molar amount (3× vector):
Insert mass:
= 7.59 × 10-13 × 1500 × 660 × 109
≈ 752 ng
Step 2: Total DNA amount:
Step 3: Calculate ligase units (using adjustment factor 2 for efficiency):
Step 4: Since 25 Units is high for a 20 µL reaction, scale down or split into multiple reactions. Typically, 1–3 Units per reaction is sufficient. The researcher can dilute DNA or perform multiple ligations.
Additional Considerations for Accurate Dose Calculations
- Enzyme Stability: Enzymes lose activity over time; always use fresh aliquots and store properly.
- Batch Variability: Different lots may have slight activity differences; verify units on datasheets.
- Reaction Inhibitors: Contaminants like EDTA or phenol can inhibit enzymes; purify DNA thoroughly.
- Reaction Kinetics: Some enzymes exhibit star activity or non-specific cleavage at high concentrations; avoid overdosing.
- Buffer Compatibility: Use manufacturer-recommended buffers to maintain optimal pH and ionic strength.
- Temperature Control: Incubate reactions at precise temperatures to ensure enzyme efficiency.
Authoritative External Resources for Enzyme Dose Calculations
- New England Biolabs Enzyme Usage Guidelines – Industry standard for restriction enzyme and ligase dosing.
- Promega Ligase Activity Assay Protocol – Detailed methods for ligase quantification.
- Thermo Fisher PCR Optimization Guide – Guidelines for polymerase dosing and reaction setup.
- NCBI Article on Enzyme Kinetics in Molecular Biology – In-depth review of enzyme activity and dosing.