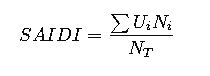Discover the essential calculation of Km and Vmax in Michaelis-Menten kinetics; our guide simplifies complex enzyme kinetics effortlessly for you.
Master determination of Michaelis-Menten parameters using step-by-step procedures, detailed examples, and practical applications empowering your scientific research for breakthrough insights.
AI-powered calculator for Calculation of Km and Vmax (Michaelis-Menten Curve)
Example Prompts
- 10, 20, 0.5
- 15, 30, 0.8
- 25, 50, 1.2
- 5, 10, 0.3
Understanding Michaelis-Menten Kinetics
The Michaelis-Menten equation is foundational in enzyme kinetics, describing how reaction velocity relates to substrate concentration. At its core, the equation offers insight into the efficiency and affinity of an enzyme toward its substrate.
Modern biochemical research relies on accurate determination of Km and Vmax, which represent, respectively, the substrate concentration at half-maximal velocity and the maximum rate achieved by the enzyme. This calculation is critical in fields ranging from drug development and clinical diagnostics to industrial biotechnology.
The equation is defined as V = (Vmax × [S]) / (Km + [S]). Here, V represents the reaction velocity at a given substrate concentration [S]. Vmax is the maximum velocity that the enzyme can achieve when fully saturated with substrate, while Km is the substrate concentration at which the reaction velocity reaches half of Vmax.
Researchers and engineers derive these parameters by collecting experimental data, plotting substrate concentration versus reaction velocity, and applying various mathematical approaches such as non-linear regression or linear transformations. These techniques have become indispensable in assessing enzyme performance.
Beyond theoretical understanding, the calculation of Km and Vmax is also implemented in advanced software and laboratory instruments. The accuracy of these values directly influences the design of enzyme inhibitors, formulation of pharmaceuticals, and overall control of biochemical processes.
The procedure for determining Km and Vmax includes precise measurement of substrate concentrations and reaction velocities under controlled conditions. Critical analysis and statistical treatment of these measurements ensure reliable parameter estimations that are essential for biochemical modeling.
Furthermore, engineers and biochemists use simulation tools to predict enzyme behavior. The integration of these parameters into dynamic models enhances the understanding of metabolic pathways and facilitates the optimization of industrial bioreactors for improved product yields.
The interplay of substrate concentration, enzyme saturation, and reaction rate is not only pivotal in academic research but also in practical applications. Industries such as biofuel production and food processing utilize these principles to maximize enzymatic efficiency and resource utilization.
Enzyme kinetics experiments often require multiple replicates and a wide range of substrate concentrations. Proper experimental design coupled with analytical rigor allows for effective determination of the Michaelis-Menten parameters, ensuring robust and reproducible results.
Basic Formulas for Calculation of Km and Vmax
The fundamental Michaelis-Menten equation is represented as:
V = (Vmax × [S]) / (Km + [S])
Here, each variable is defined as follows:
- V: Reaction velocity at a particular substrate concentration [S].
- Vmax: Maximum reaction velocity when the enzyme is saturated with substrate.
- [S]: Substrate concentration in the reaction mixture.
- Km: Michaelis constant, indicating the substrate concentration where V = Vmax / 2.
This formula is used to determine how the enzyme’s activity changes with varying substrate concentrations. A plot of V versus [S] will typically show a hyperbolic curve.
An alternative and mathematically advantageous method is the Lineweaver-Burk transformation, which linearizes the data by taking the reciprocal:
1/V = (Km/Vmax)(1/[S]) + 1/Vmax
The Lineweaver-Burk plot is generated by plotting 1/V versus 1/[S]. This linear representation allows for easy extraction of Km and Vmax from the intercepts and slope.
Another graphical method employs the Eadie-Hofstee plot where V is plotted against V/[S]. The corresponding equation is:
V = Vmax – Km(V/[S])
In this plot, the intercept equals Vmax, and the slope’s magnitude gives Km, providing an alternative approach with different error distribution properties.
Each of these methods has distinct advantages. The Michaelis-Menten direct plot offers a visual analysis, whereas the transformed plots permit straightforward linear regression. Selecting the right approach depends on data quality and experimental conditions.
Tables Summarizing Michaelis-Menten Formulas
| Formula | Description |
|---|---|
| V = (Vmax × [S]) / (Km + [S]) | Standard Michaelis-Menten equation defining the relationship between reaction velocity (V), substrate concentration ([S]), Vmax, and Km. |
| 1/V = (Km/Vmax)(1/[S]) + 1/Vmax | Lineweaver-Burk transformation that linearizes the Michaelis-Menten relationship for easier parameter extraction. |
| V = Vmax – Km(V/[S]) | Eadie-Hofstee plot rearrangement, providing another linear representation of enzyme kinetics data. |
These formulas form the backbone of enzyme kinetic analysis and serve as essential tools for experimental design in biochemistry.
The choice of graphical method depends on data consistency and the presence of outliers. It is often beneficial to analyze the same data set with multiple methods to confirm the accuracy of calculated parameters.
Advanced Calculation Methods for Km and Vmax
Beyond simple graphical methods, modern enzymology leverages computational techniques to perform non-linear regression analysis. These methods are particularly useful when data points contain experimental errors or when substrate inhibition might be present.
Software packages, such as GraphPad Prism, MATLAB, and various Python libraries, provide robust tools for fitting the Michaelis-Menten model to experimental data. By employing iterative algorithms like the Levenberg-Marquardt method, these programs can converge on the best-fit parameters for Km and Vmax with high accuracy.
Non-linear regression minimizes the sum of squared differences between experimental data points and the model’s predicted values. This approach statistically weighs each data point, thereby providing a more reliable estimate under a range of substrate concentrations.
In addition to regression analysis, error propagation techniques are used to estimate the uncertainty associated with Km and Vmax. Understanding these errors is crucial when using kinetic parameters for further research or in designing inhibitors based on enzyme efficiency.
In practical settings, data are often transformed using double reciprocal plots or Eadie-Hofstee plots to expedite preliminary analysis. Nonetheless, the gold standard remains non-linear regression for its enhanced precision.
Modern analytical instruments can automatically record reaction velocities and substrate concentrations over time. These instruments generate time-course data that, when integrated with computational tools, offer a dynamic view of enzyme kinetics under variable conditions.
Furthermore, simulation models can predict enzyme performance across a range of environmental parameters. These in silico experiments provide insights that support the design of more effective biochemical assays and industrial processes.
The integration of computational tools with experimental techniques continues to refine the accuracy of kinetic measurements, making it easier for scientists to compare data across laboratories and studies.
Graphical Analysis Techniques
Graphical methods remain a popular choice to visualize and understand the Michaelis-Menten curve. A typical V versus [S] plot produces a hyperbolic curve, which can be used to visually approximate Vmax and Km.
High-quality plots are crucial for accurate parameter estimation. Researchers are encouraged to use software that allows for proper scaling of axes and offers fitting tools to overlay theoretical curves on experimental data.
The Lineweaver-Burk plot is especially useful when linearity is required for simplicity. By plotting 1/V against 1/[S], one obtains a straight line where the y-intercept equals 1/Vmax and the x-intercept relates to -1/Km.
The Eadie-Hofstee plot, which graphs V against V/[S], has the advantage of maintaining uniform error distribution over the entire range of substrate concentrations. This can lead to better parameter estimates when the error variance is homoscedastic.
It is essential to note that each graphical method has inherent limitations. For example, Lineweaver-Burk plots can overweight lower substrate concentration data, potentially leading to inaccuracies if not handled with care.
Therefore, it is common practice to cross-validate these methods with non-linear regression analyses. Such cross-validation not only bolsters confidence in the results but also highlights any potential discrepancies in the data.
Consistency between graphical and computational methods is key to ensuring the reliability of the determined values. When discrepancies are found, they often trigger a re-examination of experimental conditions and data collection methods.
Overall, graphical analysis serves as both an educational tool and a practical method for initial evaluation, forming an indispensable part of enzyme kinetics research.
Real-World Application Case Studies
Real-world examples provide necessary context and validation for the theoretical approaches discussed. Here, we detail two in-depth application cases to showcase the practical use of Km and Vmax calculations.
Case Study 1 involves the evaluation of a novel enzyme used in drug metabolism research. The goal was to determine the kinetic parameters to understand how efficiently the enzyme could metabolize a new drug candidate.
In the laboratory, the enzyme’s reaction velocities were measured at various substrate concentrations. The raw data were as follows:
| Substrate [S] (mM) | Velocity (µmol/min) |
|---|---|
| 0.5 | 12 |
| 1.0 | 20 |
| 2.0 | 30 |
| 4.0 | 36 |
| 8.0 | 38 |
Using non-linear regression, the experimental data were fitted into the Michaelis-Menten model. This analysis yielded a calculated Vmax of approximately 40 µmol/min and a Km of 1.5 mM. These results indicated that the enzyme was highly efficient at low substrate concentrations, correlating with the predicted metabolic rate of the drug.
The significance of these findings lies in optimizing drug dosage. With precise values for Km and Vmax, pharmacologists can make informed decisions about enzyme inhibitors, drug interactions, and potential side effects. External resources such as the NCBI database provide additional validation and similar case studies supporting these conclusions.
Case Study 2 explores an industrial biotechnology application where enzyme kinetics are used to optimize an enzymatic process for biofuel production. In this scenario, an enzyme catalyzes the breakdown of biomass to generate fermentable sugars.
Experimental data were collected at different substrate concentrations. The raw data produced the following values:
| Substrate [S] (g/L) | Reaction Rate (g/L/h) |
|---|---|
| 10 | 5 |
| 20 | 9 |
| 50 | 14 |
| 100 | 16 |
| 200 | 16.5 |
Non-linear regression analysis of the biofuel production data estimated a Vmax of about 17 g/L/h and a Km of roughly 30 g/L. These parameters provided valuable insights into the enzyme’s performance and the optimal substrate range, improving efficiency in industrial-scale operations.
Industrial engineers used these values to adjust reactor conditions, maximizing yield while minimizing unintended side reactions. Detailed process control was established based on the kinetic parameters. The study information is corroborated by findings published in reputable journals such as the ACS Publications.
Additional Considerations in Enzyme Kinetics Analysis
When applying the Michaelis-Menten model, it is essential to consider factors such as enzyme stability, substrate purity, and potential inhibitors present in the reaction mixture. These variables can influence the experimental determination of Km and Vmax.
Careful control of experimental conditions is paramount. Researchers should ensure that enzyme concentrations remain constant throughout the assay and that substrate depletion does not influence reaction kinetics. Techniques such as pre-incubation and dilution protocols are frequently employed to maintain consistency across experiments.
Another important consideration is the impact of environmental conditions such as pH and temperature. Variations in these parameters can alter enzyme conformation and, subsequently, its kinetic behavior. Thus, environmental controls and calibration experiments are essential to discern true kinetic constants from experimental artifacts.
To further enhance the accuracy of kinetic measurements, advanced techniques such as stopped-flow spectroscopy and isothermal titration calorimetry may be employed. These methods offer real-time monitoring of rapid enzymatic reactions and allow for precise measurements in dynamic systems.
Modern analytical techniques, coupled with robust statistical analysis, are pushing the frontiers of enzyme kinetics research. With the advancement in computational power and data acquisition systems, researchers can now model enzyme activity under complex conditions, providing deeper insights into biochemical processes.
It is also recommended to perform replicates and independent validations to ensure reproducibility of the calculated Km and Vmax values. Peer-reviewed studies and collaborative research play a significant role in confirming the reliability of kinetic models.
Moreover, sensitivity analysis can be conducted to assess how small changes in substrate concentration or enzyme activity affect the calculated parameters. This analysis is crucial in validating the robustness of the experimental setup and the subsequent kinetic interpretations.
Frequently Asked Questions
Q1: What is the significance of Km in enzyme kinetics?
A: Km represents the substrate concentration at which the reaction velocity is half of Vmax. It is an indicator of the enzyme’s affinity for its substrate—a lower Km signifies higher affinity.
Q2: How is Vmax determined experimentally?
A: Vmax is obtained by measuring the reaction rate at increasing substrate concentrations until the enzyme becomes saturated. The value is typically estimated using non-linear regression on experimental data.
Q3: Why use a Lineweaver-Burk plot?
A: The Lineweaver-Burk plot linearizes the Michaelis-Menten equation, allowing easier extraction of Km and Vmax using linear regression. However, it may overemphasize errors at low substrate concentrations.
Q4: What software tools assist in calculating Km and Vmax?
A: Several tools such as GraphPad Prism, MATLAB, and Python libraries (like SciPy) can help perform non-linear regression analysis to accurately determine enzyme kinetic parameters.
Q5: Can other factors impact the estimation of Km and Vmax?
A: Yes, variables including enzyme stability, pH, temperature, and the presence of inhibitors may impact the accurate determination of kinetic parameters, necessitating stringent experimental controls.
Optimizing Data Analysis for Enzyme Kinetics
Achieving reliable calculations of Km and Vmax requires both high-quality data and appropriate analytical methods. Researchers must design experiments to ensure uniform substrate distribution and minimize experimental error.
It is advisable to perform a wide range of substrate concentrations to fully capture the hyperbolic curve characteristic of Michaelis-Menten kinetics. Data should be collected in multiple replicates, and the resulting averages used for regression analysis to eliminate anomalies.
Automation in data collection and analysis has become standard practice in many modern laboratories. Advanced spectrophotometric instruments and microfluidic devices now streamline the process, offering real-time feedback and improved accuracy.
Additionally, software tools often provide detailed analysis features such as confidence intervals, goodness-of-fit metrics, and sensitivity analyses, which are crucial for validating the calculated parameters. This analytical rigor supports the translation of experimental data into actionable insights for both academic and industrial applications.
Optimizing data interpretation also involves understanding the limitations of each method. For instance, while graphical methods offer visual insights, they might be influenced by outlier data points. Combining these with robust computational methods ensures that the final parameters accurately reflect the enzyme’s kinetics.
In cases where substrate inhibition or cooperative binding is observed, extended models beyond the basic Michaelis-Menten equation may be necessary. These models account for allosteric effects and dynamic changes in enzyme structure, offering a more comprehensive description of the system.
For such complex systems, advanced computational modeling, including simulation and optimization algorithms, is employed. This approach leverages high-performance computing to fine-tune experimental variables and predict kinetic behavior under diverse conditions.
Practical Recommendations for Engineers and Researchers
When calculating Km and Vmax, it is vital to plan experiments meticulously. Engineers and researchers are encouraged to follow standardized protocols and use validated instruments and software to ensure that experimental data is of the highest quality.
Some practical recommendations include:
- Use a broad range of substrate concentrations to capture the full kinetic profile.
- Perform multiple replicates to ensure statistical reliability.
- Employ both graphical and non-linear regression methods for cross-validation of results.
- Control environmental factors such as pH, temperature, and ionic strength meticulously.
- Incorporate error analysis and sensitivity studies to establish the robustness of the results.
Implementing these best practices not only improves the accuracy of Km and Vmax calculations but also boosts confidence during data interpretation and subsequent application in product development or clinical research.
Furthermore, collaboration with statistical experts can add value to the kinetic analysis process. Data interpretation errors can be minimized by incorporating advanced statistical models that account for experimental variability.
Continuous training and updates in the latest analytical methodologies are recommended. Researchers should attend workshops, read the latest literature, and engage in peer discussions to stay abreast of developments in enzyme kinetics.
Lastly, documentation of experimental protocols and detailed reporting of data analysis steps is critical. This transparency not only facilitates reproducibility but also helps in identifying any methodological biases or systematic errors.
Leveraging Computational Tools and External Resources
In today’s data-driven scientific landscape, computational tools play a crucial role in analyzing and interpreting enzyme kinetics data. Integration of software like GraphPad Prism, MATLAB, Python’s SciPy, and R offers seamless data fitting and error minimization.
These tools facilitate quick adjustments to models, allowing researchers to test multiple kinetic scenarios efficiently. For example, a Python script using curve_fit from the SciPy package can streamline the non-linear regression process and yield rapid estimates of Km and Vmax.
In addition, reputable external resources such as the NCBI and Sigma-Aldrich provide valuable databases, research articles, and protocols that help in refining experimental designs.
Researchers are encouraged to leverage these external links to access peer-reviewed materials and validated methodologies. Such external validation enhances the credibility and reproducibility of enzyme kinetics studies.
The integration of experimental data with computational analysis represents the future of enzyme kinetics research. As computational power increases and data acquisition becomes more streamlined, the precision of Km and Vmax determination will continue to improve, fostering advances in biochemistry and engineering.
Final Remarks
The calculation of Km and Vmax via the Michaelis-Menten model remains a cornerstone in enzymology, bridging theoretical biochemistry and practical industrial applications. From drug metabolism studies to biofuel production, accurate kinetic measurements are critical for process optimization and innovation.
By combining robust experimental design with advanced computational analysis, researchers can obtain precise estimates of these fundamental parameters. The methods discussed, including direct plotting, Lineweaver-Burk and Eadie-Hofstee transformations, and non-linear regression approaches, each offer unique insights and practical advantages.
Continuous advancements in instrumentation and data processing are pushing the boundaries of traditional enzyme kinetics. In challenging experimental scenarios where substrate inhibition or cooperative behavior is observed, extended models and simulation tools ensure that kinetic analysis remains comprehensive and accurate.
Understanding and calculating Km and Vmax not only provide critical insights into enzyme behavior but also empower researchers and engineers to design better drugs, optimize industrial processes, and develop innovative biochemical technologies. Armed with these tools and methodologies, scientist and engineers are better equipped to address complex challenges in biochemical research and industrial applications.
As the field of enzyme kinetics evolves, the integration of experimental data, computational analysis, and rigorous statistical methods will continue to be vital. Staying updated with the latest research through external links, collaborative studies, and advanced training will ensure that the determination of these kinetic parameters remains at the forefront of scientific inquiry.
This guide serves as an extensive resource for anyone involved in the calculation of Km and Vmax, offering detailed insights, step-by-step methodologies, and real-world examples that address both academic and applied challenges in enzyme kinetics.
For continued learning and further exploration of enzyme kinetics, researchers are encouraged to dive into available literature, participate in professional networks, and make use of robust computational tools designed to accurately calculate Michaelis-Menten parameters.